 |
 |
- Search
| Precis Future Med > Volume 4(1); 2020 > Article |
|
Abstract
Purpose
The purpose of this study was to investigate whether the risk of schizophrenia is causally associated with the risk of breast cancer.
Methods
Bidirectional two-sample Mendelian randomization (TSMR) was performed using genome-wide association study summary data from a total of 311,266 individuals. The causal relationship was estimated using an inverse-variance weighted method (IVW). For sensitivity analysis, weighted median, Mendelian randomization (MR)-Egger regression, and Mendelian randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) were used.
Results
The study showed evidence of a causal association between schizophrenia and the risk of breast cancer. In the IVW analysis, the odds ratio for breast cancer per log odds increase in schizophrenia risk was 1.069 with a 95% confidence interval of 1.028 to 1.112 (P< 0.001). This liability was consistently observed using multiple sensitivity analyses (P< 0.05). By contrast, the TSMR analysis of an inverted direction revealed no significant impact of breast cancer on the risk of schizophrenia.
Associations between physical illness and psychiatric disorders have sparked much interest lately. A recent meta-analysis study showed thatthe incidence of breast cancer(BC) was higher in women with schizophrenia than in healthy women [1]. However, the association was confounded by the inherent limitations of the observational studies, which included environmental factors such as psychotropic medication, hyperprolactinemia, and unhealthy lifestyles. A previous two-sample Mendelian randomization (TSMR) study showed modestly increased risk of BC in genetically prone schizophrenic women [2]. However, this study did not elucidate whether genetically determined BC risk was associated with schizophrenia. We hypothesized that the risk of schizophrenia may be causally associated with the risk of BC. To provide greater clarity in causality and biological interpretation, we conducted a bidirectional TSMR analysis to examine the causal association between schizophrenia and BC in both directions and a gene set enrichment analysis of actual genetic variants that might contribute to the association to investigate potential mechanisms [3,4].
Mendelian randomization (MR) leverages genetic variants to infer causality of the exposure-outcome association [5]. In this study, we performed TSMR by using summary statistics of the largest genome-wide association studies (GWAS) carried out on schizophrenia (n=82,315) for European ancestry and BC (n=228,951) for women of European ancestry, which were obtained from the Psychiatric Genomics Consortium (PGC) and the Breast Cancer Association Consortium (BCAC), respectively [6,7]. The TSMR analysis was performed by using MR-Base (http://www.mrbase.org/) software [8]. The inverse-variance weighted method (IVW) was used for TSMR analysis because it gives reliable combined estimates of the effect of the exposure on the outcomes, under conditions of heterogeneity between instrumental variables (IVs) that may arise when a large number of IVs are included in an MR analysis. The result of the IVW method using an IVW meta-analysis is the average ofthe ratio estimates across all genetic variants and indicates the global casual estimates [9]. The IVW method is appropriate only when all genetic variants used as IVs are valid. However, as all IVs may not satisfy the IV assumptions of MR, we performed sensitivity analyses using three additional methods, namely, weighted median (WM), MR-Egger regression, and IVW after removing pleiotropic variants using Mendelian randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) (Table 1) [10,11]. The WM gives constant estimates if more than 50% of the weight comes from valid IVs [12]. MR-Eggerregression is the same as the IVW except that it does not constrain the intercept representing the horizontal pleiotropy to zero. MR-PRESSO provides causal estimates after detecting and removing genetic variants that have horizontal pleiotropic effects in the conventional MR method (IVW). Further, we included more genetic variants in TSMR analysis based on several relaxed thresholds for the GWAS results. Leave-one-out analyses were also performed to test whether a certain variant substantially affected the causal estimate while analyzing the association between occurrence of schizophrenia and BC.
To elucidate the potential relationship between schizophrenia and BC, we performed gene set enrichment testing for evaluating the biological background shared between schizophrenia and BC using Functional Mapping and Annotation of GWAS (FUMA GWAS) [4]. Based on GWAS summary statistics of each trait, FUMA selected independent genome-wide significant single-nucleotide polymorphisms (SNPs) and their neighboring significant SNPs with low linkage disequilibrium (r2 <0.1) based on the 1,000 Genomes European data. These selected SNPs were then mapped to genes according to the functional annotation as follows: (1) positional mapping for exonic and splicing SNPs; (2) expression quantitative traitloci (eQTL) mapping based on theGenotype-Tissue Expression (GTEx) V8 eQTLs; and (3) chromatin interaction mapping using the PsychENCODE database. Using the gene set enrichment test by FUMA GWAS, we evaluated whetherthe mapped genes were over-represented in particular biological pathways. Next, we selected significantly enriched gene sets from both schizophrenia and BC (Benjamini-Hochberg false discovery rate [FDR] <0.05).
This study was approved by the Institutional Review Board of Samsung Medical Center (IRB no. SMC 2016-12-064). Informed consent was waived by the board.
Our results from the IVW and WM methods showed that schizophrenia was associated with the risk of BC (odds ratio [OR] per log odds increase in schizophrenia risk, 1.069; 95% confidence interval [CI], 1.028 to 1.112; P<0.001 forIVW) (OR, 1.038, 95% CI, 1.004 to 1.073; P<0.05 for WM) (Table 1, Fig. 1A). Further analyses across multiple GWAS thresholds showed a more significant association, possibly because of the increased statistical power (Table 1). The MR-Egger regression approach is robust, particularly for pleiotropic effects, and it revealed a moderately significant association between schizophrenia and BC. However, no significant pleiotropy was observed in the MR-Egger pleiotropy test (P> 0.05). Furthermore, the liability was consistent upon the removal of outlier variants with potential pleiotropic effects. In the MR-PRESSO test, we excluded five SNPs (rs13217619, rs1501357, rs2905426, rs4702, and rs58120505) from analysis. Significantly, the directional pleiotropy in the IVW method remained significant after removing the outliers (OR, 1.041; 95% CI, 1.013 to 1.070; P<0.005). Although we observed moderate heterogeneity between IVs (heterogeneity I2=70%) (Table 1, Fig. 1B), our results from leave-one-out analysis confirmed that the overall IVW estimates were not affected significantly by each variant (Fig. 2). The results for funnel plot asymmetry showed that the estimates on the precision (1/standard error) and Wald ratios for each SNP were bilaterally symmetrical (Fig. 1C), thereby confirming thatthe SNPs used in this analysis were valid instruments.
Next,to understand the potential mechanism ofthe causal association, we tested whether genes associated with the instrumental genetic variants were enriched in specific biological pathways. Our results indicated that fourteen canonical pathways, including estrogen-dependent gene expression, DNA repair, and estrogen-mediated signaling, were significantly shared (Benjamini-Hochberg FDR <0.05) in both schizophrenia and BC (Table 2). No significant association was observed when the TSMR analysis was performed in an inverted direction, indicating that BC had no significant effect on schizophrenia risk (P>0.05).
Earlier studies have reported an association between schizophrenia and BC [1]; it is still unclear whether the higher risk of BC in schizophrenic patients is attributed to schizophrenia orto other environmental factors. In observational studies, it is challenging to control for potential confounders, such as obesity, nulliparity, unhealthy lifestyles, or hyperprolactinemia associated with antipsychotic medication. In addition, so far, no randomized controlled trials have been performed in this area. MR analysis has emerged as a promising approach to examine causal associations [3]. Since allele segregation occurs randomly and is not affected by environmental confounding factors, MR analysis avoids the major limitations of conventional observational studies [13]. Our results based on bidirectional TSMR analysis showed that schizophrenia and BC might have a shared biological background. In addition, our MR analysis implied that the liability related to schizophrenia may show causality for the risk of BC. Our results were similar to those showing increased risk in a previous epidemiological study [14]. Significantly, our findings are consistent with a recent report suggesting a positive association between genetically determined schizophrenia and the risk of ER-positive as well as ER-negative BC [2].
Sensitivity analyses of multiple TSMR methods and multiple GWAS P-value cut-offs ensured the reliability of the causal association between schizophrenia and BC presented in this study. In the presence of pleiotropic effects, MR-Egger regression provided a robust causal estimate of instrumental genetic variants. In contrast, IWV and WM demonstrated increased power for the causal inference under the “no-pleiotropy” assumption. Ourresults from both, IWV and WM data, showed a significant causal association between schizophrenia and BC. Moreover, the inclusion of MR-Egger and MR-PRESSO methods ensured that our analysis was unlikely to violate the MR assumptions.
The association between schizophrenia and cancerrisk remains poorly understood [1]. Some of the previous studies reported a reduced risk of lung cancer in patients with schizophrenia or melanoma [15]. In addition,reduced overall cancer risk was reported in first-degree relatives of patients with schizophrenia [16]. In light of available scientific literature, and our findings, it is clear that the relationship between schizophrenia and cancers may be complicated and vary across cancer types. Besides the confounding factors, decreased life expectancy in patients with schizophrenia may also obscure its relationship with BC, which commonly afflicts women after middle age.
Ourresults from FUMA GWAS showed thatthe genes ofthe instrumental genetic variants were enriched in canonical pathways, such as estrogen-dependent gene expression, DNA repair, and estrogen-mediated signaling. Estrogen is catabolized by hydroxylation reactions, which is related to catechol-O-methyltransferase (COMT) by catechol homeostasis. The low activity of COMT has also been reported to be related to aggressive behavior in schizophrenia, and increased risk of BC [17]. Moreover, the dysregulated DNA repair canonical pathway has been indicated in BC [18] and schizophrenia [19]. These results supportthat BC and schizophrenia may have a shared genetic background, and explain the causalrelationship observed between schizophrenia and BC in our study. The underlying mechanism of the association between schizophrenia and BC needs to be investigated in further studies.
Multiple diseases of the central nervous system, such as Down’s syndrome, Parkinson’s disease, Alzheimer’s disease, anorexia nervosa, and multiple sclerosis are positively or negatively associated with the risk of cancer [20]. Emerging reports have shown that the immune and nervous systems might play a part in the development of malignancy [21]. However, further research is warranted to validate these observations.
Although our results provide reliable evidence of the increased risk of BC in schizophrenia patients,there are several limitations in this study. First, since binary exposure (schizophrenia) was tested in the present TSMR analysis, we acknowledge that there may be a continuous latent variable related to the psychological problem for the dichotomized schizophrenia [22]. MR association between binary exposure and binary outcome can only infer shared liability between the two conditions. Our result may be interpreted to suggest that the risk factors of schizophrenia may also raise the risk of BC. Second, the GWAS sample size for schizophrenia was much smaller than the GWAS sample size for BC. Although our results indicated that the liability from schizophrenia to BC was significant, further analysis of GWAS results based on a comparable sample size may be necessary to validate these results.
Our results suggest that concrete efforts are required to mitigate the burden of malignant diseases in schizophrenia patients. For instance, the development of breast screening guidelines in schizophrenia patients might be helpful for the early diagnosis of disease in these patients. On the flip side, early screening and preventive measures could also lead to adverse consequences, such as overdiagnosis, false-positive findings, patient anxiety, unnecessary treatment and associated risks, and poor compliance to psychotropic medications [23]. Therefore,the development of a BC risk model for schizophrenia patients that include other known risk factors is essential for decision-making regarding the intensity of surveillance. A multidisciplinary research consortium comprising researchers from multiple specialists including psychiatrists, oncologists, and geneticists might be constituted to address this issue.
In conclusion, our results showed a potential clinical relevance between schizophrenia and BC. Our findings suggest the need for further clinical consideration of BC screenings and studies to identify the biological mechanisms underlying the potential causal association between schizophrenia and BC to promote health and quality of life in patients with schizophrenia.
Notes
AUTHOR CONTRIBUTIONS
Conception or design: SK, KK, WM, HHW.
Acquisition, analysis, or interpretation of data: SK, KK, WM, HL, HK, DKK, HHW.
Drafting the work orrevising: SK, KK, WM, HHW.
Final approval ofthe manuscript: WM, HHW.
ACKNOWLEDGEMENTS
This research was supported by the National Research Foundation of Korea (NRF) grantfunded by the Ministry of Science and ICT (Information and Communication Technologies) of Korea government (NRF-2019R1A2C4070496, Hong-Hee Won; and NRF-2018R1C1B6001708, Woojae Myung).
Fig. 1.
Estimated positive causal effects of genetically increased schizophrenia risk on breast cancer risk. (A) Scatter plot of the effect size for risk of schizophrenia and breast cancer per genetic variant. (B) Forest plot representing the causal estimation of schizophrenia risk on breast cancer risk using each or all variants. (C) Funnel plot showing symmetry that indicates the absence of heterogeneity due to horizontal pleiotropy. IVW, inverse-variance weighted method; MR, Mendelian randomization; MR-PRESSO, Mendelian randomization Pleiotropy RESidual Sum and Outlier; OR, odds ratio; SE, standard error.
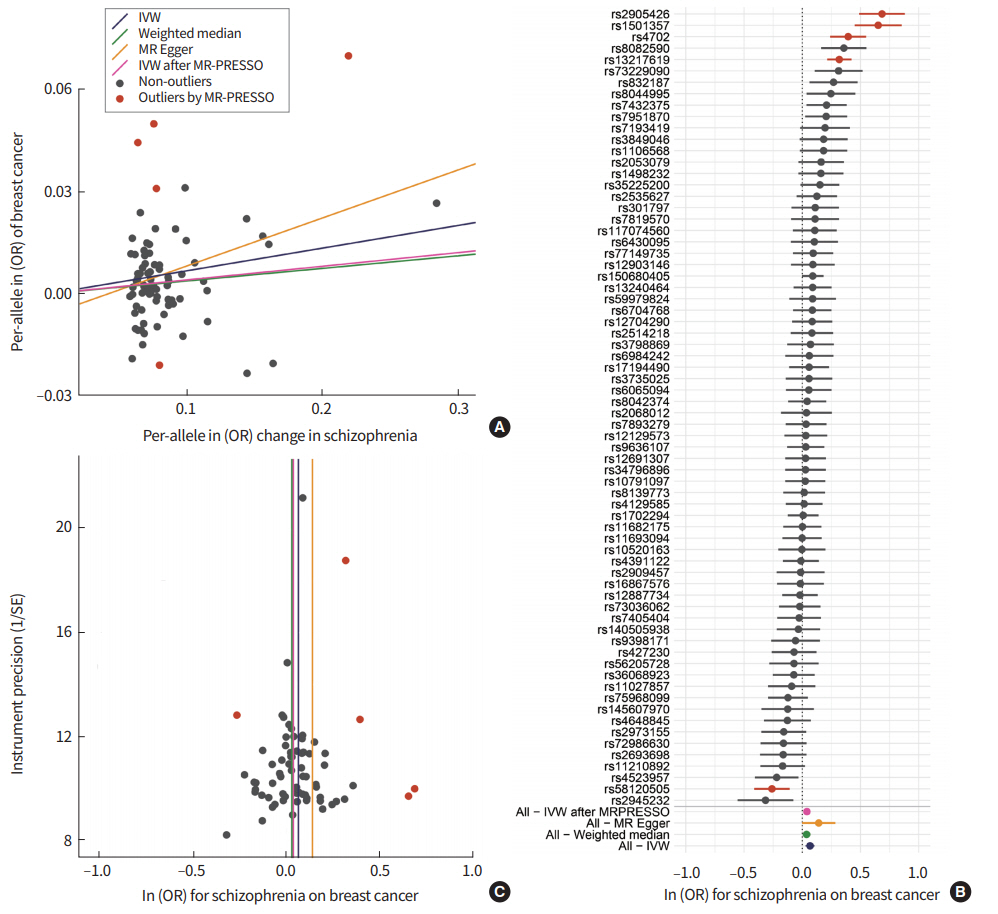
Fig. 2.
Leave-one-out sensitivity analysis for the causal effect of schizophrenia on the risk for breast cancer. OR, odds ratio.
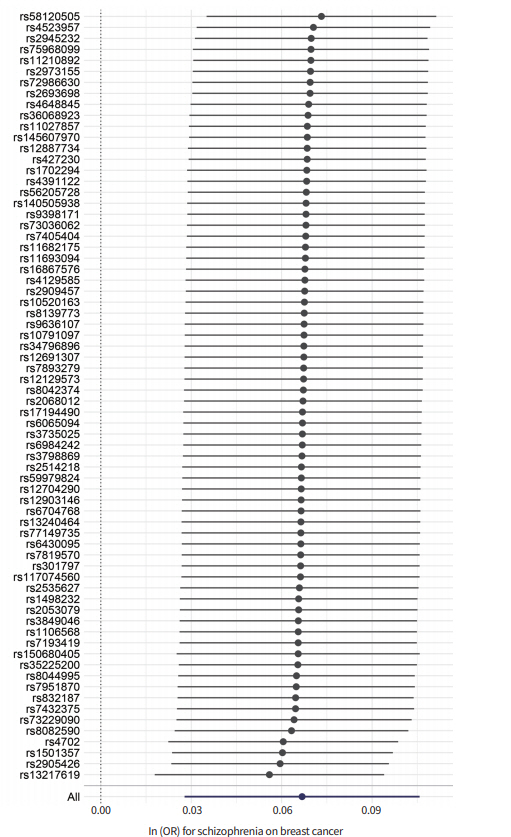
Table 1.
Two-sample MR results of schizophrenia and breast cancer using multiple MR methods
MR, Mendelian randomization; GWAS, genome-wide association study; IVW, inverse-variance weighted method; SNP, single-nucleotide polymorphism; OR, odds ratio for breast cancer; CI, confidence interval; Q, Cochran's heterogeneity statistic; Qdf, the degrees of freedom; I2, 100%×(Q–Qdf)/Q; SE, standard error; MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier.
Table 2.
Enrichment of the significant genes selected from schizophrenia (SCZ) and breast cancer (BC) GWAS in canonical pathways
REFERENCES
1. Zhuo C, Triplett PT. Association of schizophrenia with the risk of breast cancer incidence: a meta-analysis. JAMA Psychiatry 2018;75:363–9.



2. Shi J, Wu L, Zheng W, Wen W, Wang S, Shu X, et al. Genetic evidence for the association between schizophrenia and breast cancer. J Psychiatr Brain Sci 2018;3:7.


3. Byrne EM, Yang J, Wray NR. Inference in psychiatry via 2-sample Mendelian randomization-from association to causal pathway? JAMA Psychiatry 2017;74:1191–2.


4. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826.




5. Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet 2018;19:110–24.



6. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7.




7. Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancerrisk loci. Nature 2017;551:92–4.




8. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408.




9. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–89.




10. Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep 2017;4:330–45.




11. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–8.




12. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14.



13. Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet 2015;16:327–50.


14. Irwin KE, Park ER, Shin JA, Fields LE, Jacobs JM, Greer JA, et al. Predictors of disruptions in breast cancer care for individuals with schizophrenia. Oncologist 2017;22:1374–82.



15. Catala-Lopez F, Suarez-Pinilla M, Suarez-Pinilla P, Valderas JM, Gomez-Beneyto M, Martinez S, et al. Inverse and direct cancer comorbidity in people with central nervous system disorders: a meta-analysis of cancer incidence in 577,013 participants of 50 observational studies. Psychother Psychosom 2014;83:89–105.


16. Catts VS, Catts SV, O’Toole BI, Frost AD. Cancer incidence in patients with schizophrenia and their first-degree relatives: a meta-analysis. Acta Psychiatr Scand 2008;117:323–36.


17. Volavka J, Czobor P, Cseh A, Lachman H, Saito T, et al. Suicidal behavior in patients with schizophrenia is related to COMT polymorphism. Psychiatr Genet 2000;10:117–24.


18. Hussain S, Saxena S, Shrivastava S, Mohanty AK, Kumar S, Singh RJ, et al. Gene expression profiling of spontaneously occurring canine mammary tumours: insight into gene networks and pathways linked to cancer pathogenesis. PLoS One 2018;13:e0208656.



19. Kerns D, Barley K, Dracheva S, Katsel P, Casaccia P, et al. Gene expression abnormalities and oligodendrocyte deficits in the internal capsule in schizophrenia. Schizophr Res 2010;120:150–8.


20. Tabares-Seisdedos R, Dumont N, Baudot A, Valderas JM, Climent J, Valencia A, et al. No paradox, no progress: inverse cancer comorbidity in people with other complex diseases. Lancet Oncol 2011;12:604–8.


21. Ondicova K, Mravec B. Role of nervous system in cancer aetiopathogenesis. Lancet Oncol 2010;11:596–601.









