 |
 |
- Search
| Precis Future Med > Volume 5(3); 2021 > Article |
|
Abstract
Purpose
Target biopsy is usually performed in Prostate Imaging Reporting and Data System (PI-RADS) 4. Still, it is unclear if adding systematic biopsy to target biopsy influences cancer detection. The aim was to assess the role of systematic biopsy for detecting significant cancer after PI-RADS 4 is targeted.
Methods
Between March 2014 and November 2018, 182 men with PI-RADS 4 underwent transrectal ultrasound (TRUS)-guided biopsy. Systematic biopsy was added to target biopsy in 128 men (Group I) by May 2018 because PI-RADS 4 was not completely visible on TRUS, while it was done in 54 men (Group II) from June 2018 regardless of lesion visibility. Significant cancer detection rates (CDRs) were compared between the groups regarding target and systematic biopsies. Major complication rate was also compared. Significant cancer was defined as a Gleason score ≥7 tumor. Standard reference was biopsy examination. Fisher’s exact were used for statistical analysis.
Results
The significant CDRs were 21.9% (28/128) in the Group I and 38.9% (21/54) in the Group II (P= 0.0273). The significant cancers of Group I and II were missed in two (1.6%) and in one (1.9%) by target biopsy, respectively. Major complication rates of these groups were 0.8% (1/128) and 0% (0/54), respectively (P= 0.999).
Prostate Imaging Reporting and Data System version 2 (PI-RADSv2) has become widely used in the clinical practice since 2015 [1-3]. PI-RADSv2 categorizes prostate cancer based on magnetic resonance imaging (MRI) features and many studies have validated that PI-RADSv2 are useful in detecting significant cancer [4-9]. Prostate Imaging Reporting and Data System (PI-RADS) 4 is likely to have significant cancer and requires a biopsy [1-3]. Reportedly, significant cancer detection rates (CDRs) are so wide ranging from 22.1% to 78.0% [4-6,8,10]. Mainly target biopsy has been performed because PI-RADS 4 has so strong signal intensity on MRI and is likely to have significant cancer. Because the other prostate is mainly PI-RADS 1 or 2, it is still unclear if systematic biopsy following target biopsy influences significant detection. In these categories, significant cancer is highly unlikely or unlikely to be present [1-3]. However, it does not imply that PI-RADS 1 or 2 has 0% significant cancer rate even if the significant CDR is relatively low.
PI-RADS 5 is larger and has much higher likelihood of significant cancer. Accordingly, this category hardly needs to have systematic biopsy. In contrast, PI-RADS 3 has much lower likelihood of significant cancer, so that systematic biopsy is essential to improve CDR. As a result, we have a question “Is systematic biopsy necessary to detect significant cancer in patients with PI-RADS 4?”
The correlation between systematic biopsy and CDR is very few reported in men with PI-RADS 4. Several investigators have reported that systematic biopsy following target biopsy contributes to detecting additional significant cancers [11,12]. One of the reasons why systematic cores should always be added is targeting error. It is much easier to miss a small lesion in big prostates. Falagario et al. [13] demonstrated that MRI-suspicion lesion volume percentage and lesion location are significant predictors of systematic biopsy in multivariable analysis. However, they did not demonstrate the utility of concurrent systematic biopsy in men with PIRADS 4 alone. Our hypothesis was that adding systematic biopsy to target biopsy contributes to detecting significant cancer in these patients. The purpose of our study was to assess the role of systematic biopsy for detecting significant cancer after PI-RADS 4 is targeted.
This retrospective study was approved by our Institutional Review Board of Samsung Medical Center (2019-06-089) and informed consent was waived.
Between March 2014 and November 2018, 850 men underwent prostate biopsy (Fig. 1). Of these patients, 668 were excluded because of the following criteria: PI-RADS 1–3 and 5 (n= 464), target biopsy alone in PI-RADS 4 (n= 156), low (< 2.5 ng/mL) prostate specific antigen (PSA; n= 23), previous treatments (n= 14), absent pre-biopsy MRI (n= 5), systematic biopsy alone in PI-RADS 4 (n= 2), transperineal biopsy (n= 2), poor quality of MRI images (n = 1), and metastatic prostate cancer (n= 1).
The remaining 182 men (median, 65.5 years [range, 40 to 83]) met for the following inclusion criteria: high (≥2.5 ng/ mL) PSA, pre-biopsy MRI, PI-RADS 4, and target and systematic biopsies. Their PSA, prostate volume, PSA density, MRI-biopsy interval, and tumor size ranged 2.5 to 30.3 ng/mL (median, 4.8 ng/mL), 12 to 193 mL (40 mL), 0.04 to 0.76 ng/ mL2 (0.12 ng/mL2), 0 to 646 days (53 days), and 2.0 to 14.6 mm (8.4 mm), respectively. These patients were divided into Group I (n= 128) and II (n= 54). Biopsies in the Group I and II were conducted from March 2014 to May 2018 and from June 2018 to November 2018, respectively. The difference of Group I and II was how to perform systematic biopsy. Group I underwent systematic biopsy only in patients with a PI-RADS 4 lesion which was not clearly visible on transrectal ultrasound (TRUS). Target biopsy alone was performed in patients with a PI-RADS 4 lesion which was clearly visible on TRUS. Group II underwent systematic biopsy in all patients with a PI-RADS 4 lesion which was regardless of lesion depiction on TRUS. “Clearly visible on TRUS” indicated that we were able to identify the entire tumor margin on TRUS.
Of 182 men, 162 were biopsy-naïve patients and the remaining 20 had a history of previous biopsy. Of 20 men who had undergone previous negative biopsy results, Group I and II had 13 and 7, respectively. Patient demographics in each subgroup were summarized in the Table 1.
Multi-parametric MRI was performed prior to prostate biopsy 3T unit (Intera Achieva, Philips Healthcare, The Netherlands) and the MRI sequences included T2-weighted, diffusionweighted, and dynamic contrast-enhanced axial magnetic resonance (MR) images. A single radiologist, who had 17-year experience of genitourinary imaging and intervention, evaluated MR images and categorized PI-RADS 4 as an index lesion. All biopsies were performed by himself.
For Group I, systematic biopsy was added to target biopsy from March 2014 to May 2018 when PI-RADS 4 was not completely visible on TRUS. For Group II, it was added regardless of lesion visibility from June 2018 to November 2018. Determining whether or not systematic biopsy was necessary simply depended on how TRUS operator subjectively assessed the lesion visibility on TRUS. First, target cores were obtained within the PI-RADS 4. Next, systematic cores were obtained from the other prostate tissue categorized as PI-RADS 1 or 2. The number of target and systematic cores were not determined based on lesion volume, but operator’s experience. The biopsy operator was already familiar with the TRUS techniques and imaging features which were reported by Park and Park [14].
MRI images scanned before 2015 were uploaded to our Picture Archiving and Communication System (PACS) and reviewed to categorize prostate lesions with PI-RADSv2. Since 2015, when PI-RADSv2 was released, the original PI-RADS scores made prior to biopsy were used for analysis.
Overall CDR was calculated by the number of all cancer cases divided by the number of all biopsy cases in each group or each biopsy. Significant cancer was defined as a Gleason score (GS) ≥7 tumor. Significant CDR was calculated by the number of all GS ≥7 cases divided by the number of all biopsy cases in each group or each biopsy. Overall and significant CDRs were compared between the groups or between the biopsies.
Positive core rate (PCR) was calculated by the number of all cancer cores divided by the number of all biopsy cores. PCRs were compared between the groups or between the biopsies.
The size of peripheral PI-RADS 4 was measured at diffusion-weighted axial images and that of transition PI-RADS 4 was measured at T2-weighted axial MR images. When PI-RADS 4 involved both zones, the tumor size was measured depending on the dominant location of PI-RADS 4. When a tumor involved peripheral zone more dominantly than transition zone, the lesion size was measured on diffusion-weighted MR images.
The median numbers of total or cancer cores were compared between the groups or between the biopsies. The median GSs were compared between the groups or between the biopsies. The median GSs of the target biopsy were compared between the groups.
Major complications requiring hospitalization were recorded and the complication rate was calculated to know how many men were transferred to the emergency room due to adding systematic biopsy.
For statistical analysis, Mann-Whitney test was used to compare patient age, PSA, prostate volume, PSA density, MRI-biopsy interval, tumor size, biopsy core, and GS between the groups because all factors did not follow Gaussian distribution. Fisher’s Exact test was used to compare CDRs between the groups or the biopsies. Chi-square test was used to compare PCRs between the groups or the biopsies. Odds ratio was obtained to quantify the strength of the association between the biopsies in increasing the number of systematic biopsies. The approximation of Woolf was used to calculate 95% confidence interval. Commercially available software PASW Statistics version 20.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. A two-side P-value of less than 0.05 was considered statistically significant.
Group I had higher prostate volume than Group II (P=0.0280) (Table 1). There was no difference between the groups in terms of age (P =0.6243), PSA (P =0.9693), PSA density (P = 0.0649), MRI-biopsy interval (P=0.0558), and tumor size (P=0.5318). Of 128 men with a PI-RADS 4 in the Group I, peripheral lesions and transition lesions were 125 and 3, respectively. Of 53 cancer-proven men in the Group I, peripheral cancers and transition cancers were 50 and 3, respectively. Of 54 men with a PI-RADS 4 in the Group II, peripheral lesions and transition lesions were 51 and 3, respectively. Of 29 cancer-proven men in the Group II, peripheral cancers and transition cancers were 26 and 3, respectively. There was no difference regarding the lesion ratio (P=0.3637) or cancer ratio (P=0.6605) of peripheral and transition zones. The median number of target cores was 6 (2 to 10) in the Group I and 6 (2 to 11) in the Group II (P=0.8485). The median number of cancer cores was 4 (0 to 8) in the Group I and 5 (0 to 6) in the Group II (P=0.3263). The median GS was 6 (6 to 9) in the Group I and 7 (6 to 8) in the Group II (P=0.1006).
The overall CDRs were 41.4% (53/128) in the Group I and 53.7% (29/54) in the Group II (Table 2) (P= 0.1441). The significant CDRs were 21.9% (28/128) in the Group I and 38.9% (21/54) in the Group II (P = 0.0273). The PCRs were 15.0% (243/1,622) in the Group I and 22.4% (149/664) in the Group II (P< 0.0001). The percentage of systematic biopsy were 45.9% (128/279) in the Group I and 91.5% (54/59) in the Group II (P< 0.0001).
The overall CDRs of the target biopsy were 37.5% (48/128) in the Group I and 48.1% (26/54) in the Group II (P=0.1907) (Table 3). The significant CDRs of the target biopsy were 20.3% (26/128) in the Group I and 37.0% (20/54) in the Group II (P=0.0246). The PCRs of the target biopsy were 27.7% (191/689) in the Group I and 37.3% (120/322) in the Group II (P=0.0028).
The overall CDRs of the systematic biopsy were 25.0% (32/128) in the Group I and 31.5% (17/54) in the Group II (P=0.3674) (Table 4). The significant CDRs of the systematic biopsy were 7.0% (9/128) in the Group I and 13.0% (7/54) in the Group II (P= 0.2511). The PCRs of the systematic biopsy were 5.6% (52/933) in the Group I and 8.5% (29/342) in the Group II (P= 0.0792).
In the Group I, the overall CDR of the target biopsy was 37.5% (48/128) and that of systematic biopsy was 25.0% (32/128) (P= 0.0428) (Table 3). The significant CDR of the target biopsy was 20.3% (26/128) and that of systematic biopsy was 7.0% (9/128) (P= 0.0032). The PCR of the target biopsy was 27.7% (191/689) and that of combination biopsy was 5.6% (52/933) (P< 0.0001).
In the Group II, the overall CDR of the target biopsy was 48.1% (26/54) and that of systematic biopsy was 31.5% (17/54) (P= 0.1153) (Table 4). The significant CDR of the target biopsy was 37.0% (20/54) and that of the systematic biopsy was 13.0% (7/54) (P= 0.0070). The PCR of the target biopsy was 37.3% (120/322) and that of systematic biopsy was 8.5% (29/342) (P< 0.0001).
Of 53 cancer-proven cases in the Group I, 27 were diagnosed by both, 21 by target biopsy, and five by systematic biopsy. The cancers (n=5) detected by systematic biopsy alone were all GS 6 tumors. Two significant cancers detected by systematic biopsy were GS 7 tumors but underscored as GS 6 by target biopsy.
Of 29 cancer-proven cases in the Group II, 14 were diagnosed by both, 12 by target biopsy, and three by systematic biopsy. The cancers (n=3) detected by systematic biopsy alone were all GS 6 tumors. One significant cancer detected by systematic biopsy were a GS 7 tumor but underscored as GS 6 by target biopsy (Fig. 2).
The complication rates of the groups requiring hospitalization were 0.8% (1/128) and 0% (0/54), respectively (P=1.0000). Acute prostatitis was developed in one Group I patient, but was cured by oral medication alone.
PI-RADS 4 is defined as a category with the same MR signal intensities as PI-RADS 5 [1-3]. Theoretically, the difference between the two categories is tumor size or aggressive features such extra-capsular extension or seminal vesicle invasion. PI-RADS 4 is visible on MRI as clearly as PI-RADS 5 and thus mainly target biopsy is performed under MRI-TRUS fusion or in-bore MRI guidance. Another reason for target biopsy is that significant cancer in this category is more frequent than PI-RADS 1–3 [4-9]. Our study also demonstrated that PI-RADS 4 was visible on TRUS even though the tumor margin is not completely seen.
The CDR of PI-RADS 4 is inferior to that of PI-RADS 5 [4-9]. Besides, the size of PI-RADS 4 is smaller than that of PI-RADS 5, either. Therefore, target biopsy may not be sufficient for detecting significant cancer in PI-RADS 4. Our study showed that systematic biopsy identified a significant number of prostate cancers in patients in whom PI-RADS 4 was not proven as cancer by target biopsy. In a few cases, insignificant cancer was detected in PI-RADS 4 by target biopsy whereas significant cancer was detected in the other prostate by systematic biopsy. Systematic biopsy detected significant cancer in other prostate tissue although PI-RADS 4 was not confirmed as significant cancer with target biopsy.
Several studies have reported that overall or significant CDR in PI-RADS 1 or 2 ranges from 14% to 25% [4,6,8,15]. Our study showed that the overall and significant CDRs ranged from 25.0% to 31.5% and from 7.0% to 13.0%, respectively. The significant CDRs in PI-RADS 1 or 2 are much lower than those [4-6,8,10] in PI-RADS 4 but should be concerned because systematic biopsy has potential to detect additional significant cancers. Therefore, systematic biopsy may play a great role in men with PI-RADS 4 which is not diagnosed as cancer by target biopsy. From this point of view, multifocal significant cancers are not uncommon in PI-RADS 4 [16-18]. Accordingly, before conducting a focal therapy to treat significant cancer in PI-RADS 4 unilaterally, systematic biopsy is necessary to detect additional significant cancers even though the other prostate is categorized as PI-RADS 1 or 2 on MRI.
There are several reasons that significant cancer can be missed in PI-RADS 4 by target biopsy. First, inflammation may manifest as PI-RADS 4 on MRI. Although image-guided biopsy targets it well, it turns out to be benign in men with high PSA, resulting from the other prostate categorized as PIRADS 1 or 2 [19,20]. Systematic biopsy helps to detect significant cancer in this situation. Second, PI-RADS 4 may be mistargeted. This category is measured less than 15 mm. Thus, if PI-RADS 4 is smaller and/or farther from the TRUS probe, it is difficult to target it precisely. Third, another factor to mistargeting is intrinsic in the biopsy devices. When a biopsy guideline is placed in the lesion center, a biopsy needle may travel a little off the line. Fourth, lesion location or shape influence on lesion targeting. If thin and lenticular PI-RADS 4 is attached on the anterior capsule, it is also hard to obtain pathologic tissues. A biopsy needle has a 5 mm tip which cannot contain core tissue. Therefore, the biopsy needle should penetrate the anterior capsule 5 mm to sample the lesion. Post-biopsy bleeding may increase in this situation.
Our study had some limitations. First, there were no objective criteria about when to add systematic biopsy in PI-RADS 4. Our study added it to target biopsy according to the lesion visibility. Determining the lesion visibility relied on operator’s skill and experience. Second, the optimal protocol of systematic biopsy was not assessed. The median number of systematic cores was 6 in our study. Simply increasing number of these cores improves not only significant CDR, but also increases complication rate such as pain, urinary retention, bleeding, and infection [21-24]. Third, the number of target cores was relatively high. Still, optimal cores of target biopsy are not established. Although GS 4 component exists in PIRADS 4, target biopsy only obtained from the center of PIRADS 4 may miss significant cancer if it is relatively small and eccentric. Therefore, our study sampled target cores from peripheral area as well as central area. Subsequently, the number of target cores was increased [25]. Fourth, still, TRUS-guided target biopsy is not widely accepted as a biopsy technique [26-28]. Therefore, our study demonstrated relatively low significant CDRs in patients with PI-RADS 4 [10].
Several factors are involved in the lower CDR of our study. First, our patients with PI-RADS 4 are all Asian. Generally, they had a low incidence of prostate cancer. This may influence on CDR mostly. Second factor was that Group I had a poor depiction of PI-RADS 4 on TRUS. Therefore, lesion targeting is relatively poor and systematic biopsy detected additional significant cancer. Third factor was that Group II had a good depiction of PI-RADS 4 on TRUS and thus lesion targeting is relatively better compared to Group I. Fourth factor was that 156 cases undergoing target biopsy alone were excluded. Their PI-RADS 4 lesions were clearly visible on TRUS and thus systematic biopsy was not added to target biopsy. Fifth, we did not consider PSA density, 4Kscore, and risk calculators to determine whether biopsy was necessary. These factors are useful to avoid unnecessary biopsy because negative and positive predictive values of MRI findings vary greatly based on disease prevalence [29-31]. Sixth, a single radiologist performed both MRI interpretation and prostate biopsy. Accordingly, we were not able to avoid interpretation bias because of inter-reader variability of PI-RADS score [32]. Last, our study design was retrospective.
In conclusion, systematic biopsy should be added to target biopsy even though PI-RADS 4 is clearly visible. A significant number of significant cancers are detected with systematic biopsy. Therefore, adding systematic biopsy to target biopsy may contribute to improving CDR in PI-RADS 4 because target biopsy alone may miss significant cancers.
Notes
AUTHOR CONTRIBUTIONS
Conception or design: HSK, BKP.
Acquisition, analysis, or interpretation of data: HSK, BKP.
Drafting the work or revising: HSK, BKP.
Final approval of the manuscript: HSK, BKP.
Fig. 1.
Flow diagram showing a final study population. PSA, prostate specific antigen; MRI, magnetic resonance imaging; PI-RADS, Prostate Imaging Reporting and Data System; TRUS, transrectal ultrasound.
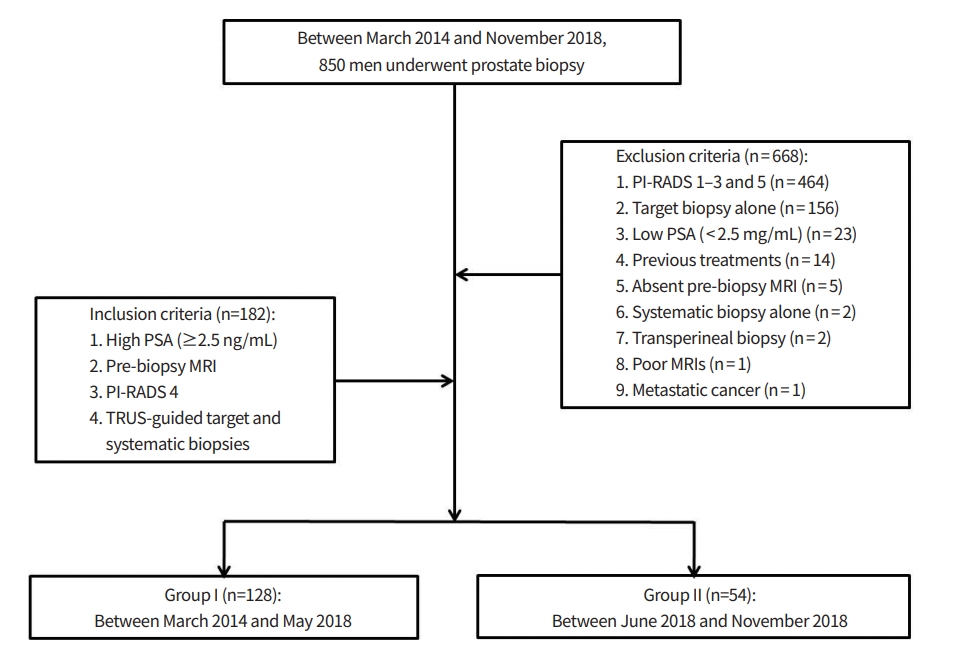
Fig. 2.
A 60-year-old man with prostate specific antigen of 7.15 ng/mL. (A) Diffusion-weighted axial image shows a Prostate Imaging Reporting and Data System (PI-RADS) 4 peripheral lesion (arrow) in the left mid-gland. It is diagnosed as an index tumor because of high signal intensity and lesion size (6.5 mm). (B) Transrectal ultrasound transverse image shows that the tumor (arrow) is detected in the left base and very hypoechoic. Target biopsy confirmed Gleason score 6 from it, while systematic biopsy sampled Gleason score 7 from the right apex, which is shown as PI-RADS 2. The prostate is not compressed by a transrectal probe.
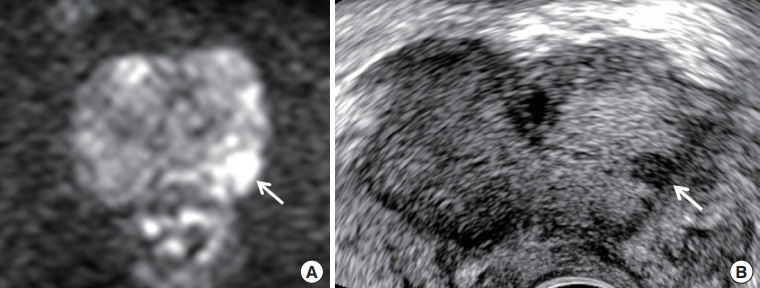
Table 1.
Patient demographics
Table 2.
Comparison of biopsy outcomes between the groups
Table 3.
Comparison of target biopsy outcomes between the groups
Table 4.
Comparison of systematic biopsy outcomes between the groups
REFERENCES
1. Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS prostate imaging: reporting and data system 2015, version 2. Eur Urol 2016;69:16–40.


2. Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, et al. Synopsis of the PI-RADS v2 Guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol 2016;69:41–9.


3. Purysko AS, Rosenkrantz AB, Barentsz JO, Weinreb JC, Macura KJ. PI-RADS version 2: a pictorial update. Radiographics 2016;36:1354–72.


4. Greer MD, Shih JH, Lay N, Barrett T, Kayat Bittencourt L, Borofsky S, et al. Validation of the dominant sequence paradigm and role of dynamic contrast-enhanced imaging in PI-RADS version 2. Radiology 2017;285:859–69.


5. Hofbauer SL, Maxeiner A, Kittner B, Heckmann R, Reimann M, Wiemer L, et al. Validation of prostate imaging reporting and data system version 2 for the detection of prostate cancer. J Urol 2018;200:767–73.


6. Mehralivand S, Bednarova S, Shih JH, Mertan FV, Gaur S, Merino MJ, et al. Prospective evaluation of PI-RADS™ Version 2 using the international society of urological pathology prostate cancer grade group system. J Urol 2017;198:583–90.



7. Park SY, Jung DC, Oh YT, Cho NH, Choi YD, Rha KH, et al. Prostate cancer: PI-RADS version 2 helps preoperatively predict clinically significant cancers. Radiology 2016;280:108–16.


8. Tan N, Lin WC, Khoshnoodi P, Asvadi NH, Yoshida J, Margolis DJ, et al. In-bore 3-T MR-guided transrectal targeted prostate biopsy: prostate imaging reporting and data system version 2-based diagnostic performance for detection of prostate cancer. Radiology 2017;283:130–9.


9. Thai JN, Narayanan HA, George AK, Siddiqui MM, Shah P, Mertan FV, et al. Validation of PI-RADS version 2 in transition zone lesions for the detection of prostate cancer. Radiology 2018;288:485–91.


10. Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med 2018;378:1767–77.



11. Ploussard G, Beauval JB, Lesourd M, Almeras C, Assoun J, Aziza R, et al. Added value of concomitant systematic and fusion targeted biopsies for grade group prediction based on radical prostatectomy final pathology on positive magnetic resonance imaging. J Urol 2019;202:1182–7.


12. Ahdoot M, Wilbur AR, Reese SE, Lebastchi AH, Mehralivand S, Gomella PT, et al. MRI-targeted, systematic, and combined biopsy for prostate cancer diagnosis. N Engl J Med 2020;382:917–28.



13. Falagario U, Jambor I, Taimen P, Syvanen KT, Kahkonen E, Merisaari H, et al. Added value of systematic biopsy in men with a clinical suspicion of prostate cancer undergoing biparametric MRI-targeted biopsy: multi-institutional external validation study. World J Urol 2020;Aug 10 [Epub]. https://doi.org/10.1007/s00345-020-03393-8.

14. Park BK, Park SY. New biopsy techniques and imaging features of transrectal ultrasound for targeting PI-RADS 4 and 5 lesions. J Clin Med 2020;9:530.



15. Cash H, Maxeiner A, Stephan C, Fischer T, Durmus T, Holzmann J, et al. The detection of significant prostate cancer is correlated with the Prostate Imaging Reporting and Data System (PI-RADS) in MRI/transrectal ultrasound fusion biopsy. World J Urol 2016;34:525–32.


16. Djavan B, Susani M, Bursa B, Basharkhah A, Simak R, Marberger M. Predictability and significance of multifocal prostate cancer in the radical prostatectomy specimen. Tech Urol 1999;5:139–42.

17. Andreoiu M, Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Hum Pathol 2010;41:781–93.


18. Wise AM, Stamey TA, McNeal JE, Clayton JL. Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology 2002;60:264–9.


19. Jyoti R, Jina NH, Haxhimolla HZ. In-gantry MRI guided prostate biopsy diagnosis of prostatitis and its relationship with PIRADS V.2 based score. J Med Imaging Radiat Oncol 2017;61:212–5.


20. Panebianco V, Giganti F, Kitzing YX, Cornud F, Campa R, De Rubeis G, et al. An update of pitfalls in prostate mpMRI: a practical approach through the lens of PI-RADS v. 2 guidelines. Insights Imaging 2018;9:87–101.


21. Nam RK, Saskin R, Lee Y, Liu Y, Law C, Klotz LH, et al. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J Urol 2010;183:963–8.


22. Loeb S, Carter HB, Berndt SI, Ricker W, Schaeffer EM. Complications after prostate biopsy: data from SEER-Medicare. J Urol 2011;186:1830–4.



23. Loeb S, Vellekoop A, Ahmed HU, Catto J, Emberton M, Nam R, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64:876–92.


24. Roberts MJ, Bennett HY, Harris PN, Holmes M, Grummet J, Naber K, et al. Prostate biopsy-related infection: a systematic review of risk factors, prevention strategies, and management approaches. Urology 2017;104:11–21.


25. Kenigsberg AP, Renson A, Rosenkrantz AB, Huang R, Wysock JS, Taneja SS, et al. Optimizing the number of cores targeted during prostate magnetic resonance imaging fusion target biopsy. Eur Urol Oncol 2018;1:418–25.


26. Park BK. Ultrasound-guided genitourinary interventions: principles and techniques. Ultrasonography 2017;36:336–48.



27. Park BK. Image-guided prostate biopsy: necessity for terminology standardization. J Ultrasound Med 2020;39:191–6.


28. Park SY, Park BK. Necessity of differentiating small (<10 mm) and large (≥10 mm) PI-RADS 4. World J Urol 2020;38:1473–9.


29. Falagario UG, Jambor I, Lantz A, Ettala O, Stabile A, Taimen P, et al. Combined use of prostate-specific antigen density and magnetic resonance imaging for prostate biopsy decision planning: a retrospective multi-institutional study using the prostate magnetic resonance imaging outcome database (PROMOD). Eur Urol Oncol 2020;Sep 21 [Epub]. https://doi.org/10.1016/j.euo.2020.08.014.

30. Falagario UG, Martini A, Wajswol E, Treacy PJ, Ratnani P, Jambor I, et al. Avoiding unnecessary magnetic resonance imaging (mri) and biopsies: negative and positive predictive value of MRI according to prostate-specific antigen density, 4Kscore and risk calculators. Eur Urol Oncol 2020;3:700–4.









